Research Projects
Recently, we and others were able to show that when transcription factors come together on DNA, they can form a liquid droplet that de-mixes from the surrounding medium, thereby transitioning from a diluted state into a separate liquid phase. Our previous work suggested that a pre-wetting-like transition is at play here (Morin et al, Nature Physics, 2022): many transcription factors bind to clusters of binding sites on the DNA that are in close proximity. This locally increases the protein concentration. As a result, the IDRs can now interact with each other strongly enough to form a condensate or liquid droplet on the DNA. Interestingly, the resulting droplets are limited in size and restricted to the DNA surface.
Our group is trying to better understand how transcriptional droplets form, which proteins they contain and how they are used to control transcriptional programs. Specifically, we use recombinant proteins to test their ability to form condensates in vitro in the presence and absence of DNA. Using single-molecule assays (Figure 1), we can probe the properties of the condensates and biophysically characterise them. By creating different mutants, we can test which parts of the protein are required for different functions or are responsible for which property. Finally, we can introduce mutations into human cells to understand how they affect transcription using state-of-the-art next-generation sequencing methods.
Figure 1: For the DNA carpet assay, the DNA is bound to a PEGylated glass cover slip, which allows visualisation of DNA molecules individually. In the next step, labelled protein is added and their condensation on the DNA can be observed in real-time. Due to the use of total internal reflection microscopy (TIRFM), single-molecule resolution can be achieved.
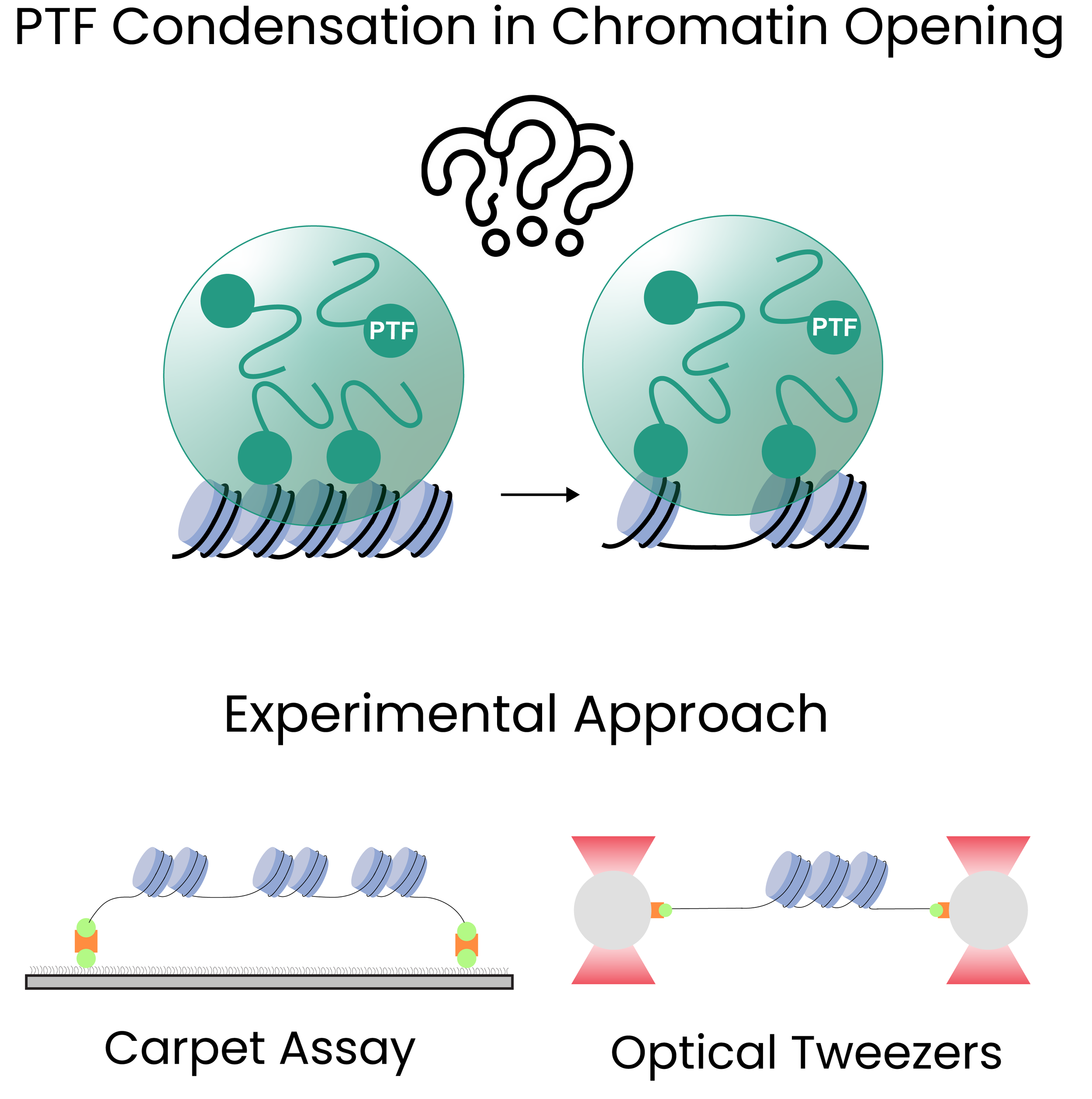
This PhD project explores how pioneer transcription factors (PTFs) use biomolecular condensation to remodel chromatin structure. While PTFs are known to access DNA within compacted chromatin, the role of their condensates in facilitating chromatin opening remains unclear. To address this, chromatin is reconstituted in vitro using fluorescently labeled histones and defined DNA templates, and its interaction with PTFs is studied using single-molecule techniques. These include optical tweezers to detect nucleosome unwrapping through force spectroscopy, and the carpet assay to visualize condensate formation on surface immobilized chromatin via TIRF microscopy. Together, these approaches allow to dissect how PTF condensation influences chromatin accessibility at the single-molecule level.
PTF Condensates in Chromatin Remodeling
The Role of Biomolecular Condensates in NGN3-Mediated Pancreatic Endocrine Differentiation
This project aims to understand whether the transcription factor (TF) Neurogenin-3 regulates pancreatic endocrine differentiation through biomolecular condensate formation. While TFs are classically understood to function via DNA-binding and co-factor recruitment, recent models highlight the role of intrinsically disordered regions (IDRs) in the formation of condensates to regulate gene expression. NGN3 contains predicted IDRs, suggesting that it may employ phase separation as a regulatory mechanism. To investigate this, we first purify recombinant WT NGN3 and IDR-targeted mutants, and assess their condensate forming potential using in vitro droplet assays. Condensate-deficient mutants will be tested for their ability to regulate cell fate in pancreatic progenitors through genomic profiling to assess transcriptional and chromatin level changes.
Together, this will help us determine how condensate formation of NGN3 contributes to its transcriptional and developmental function.
Klf4 as a Molecular Key: Unlocking Facultative Heterochromatin to Drive Cellular Identity Changes
This project investigates the reprogramming potential of Klf4, a pioneer transcription factor and one of the four canonical Yamanaka factors critical for cellular identity transitions. By introducing Klf4 into a Klf4-negative “blank slate” human cell line, the study aims to delineate its capacity to access and remodel facultative heterochromatin—regions of the genome that are developmentally poised yet remain transcriptionally silent. Through engagement with these compacted chromatin domains, Klf4 may unlock gene expression programs essential for cell fate determination.
To achieve a comprehensive understanding of Klf4’s reprogramming dynamics, the approach combines an inducible expression system with single-cell RNA sequencing, enabling precise temporal control over Klf4 activation and high-resolution mapping of transcriptional responses at the single-cell level.
A central focus of the project is to assess the impact of both native Klf4 and MBP-tagged Klf4 on the formation and regulation of transcriptional condensates—phase-separated nuclear compartments thought to organize and coordinate gene expression. Comparative transcriptomic profiling will be performed to determine how the MBP tag influences Klf4-mediated condensate formation and alters global gene expression patterns. This strategy will reveal mechanistic insights into the role of Klf4 in chromatin remodeling and transcriptional regulation, as well as potential artefacts introduced by common tagging approaches.